Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) can infect animals, however, the whole range of potential hosts is still unknown. This work makes an assessment of wildlife susceptibility to SARS-CoV-2 by analyzing the similarities of Angiotensin Converting Enzyme 2 (ACE2) and Transmembrane Protease, Serine 2 (TMPRSS2)—both recognized as receptors and protease for coronavirus spike protein—and the genetic variation of the viral protein spike in the recognition sites. The sequences from different mammals, birds, reptiles, and amphibians, and the sequence from SARS-CoV-2 S protein were obtained from the GenBank. Comparisons of aligned sequences were made by selecting amino acids residues of ACE2, TMPRSS2 and S protein; phylogenetic trees were reconstructed using the same sequences. The species susceptibility was ranked by substituting the values of amino acid residues for both proteins. Our results ranked primates at the top, but surprisingly, just below are carnivores, cetaceans and wild rodents, showing a relatively high potential risk, as opposed to lab rodents that are typically mammals at lower risk. Most of the sequences from birds, reptiles and amphibians occupied the lowest ranges in the analyses. Models and phylogenetic trees outputs showed the species that are more prone to getting infected with SARS-CoV-2. Interestingly, during this short pandemic period, a high haplotypic variation was observed in the RBD of the viral S protein, suggesting new risks for other hosts. Our findings are consistent with other published results reporting laboratory and natural infections in different species. Finally, urgent measures of wildlife monitoring are needed regarding SARS-CoV-2, as well as measures for avoiding or limiting human contact with wildlife, and precautionary measures to protect wildlife workers and researchers; monitoring disposal of waste and sewage than can potentially affect the environment, and designing protocols for dealing with the outbreak.
Severe acute respiratory syndrome coronavirus 2(SARS-CoV-2) is causing the biggest pandemic of this century, and could potentially infect between 30 and 40% of the world's populations (De Soto et al., 2020). Due to its high transmission rate and mortality, researches around the world are trying to get some insight about its origin through the analysis of related-virus genes sequences in humans and animals (Lu et al., 2020b), and looking for Angiotensin-Converting Enzyme 2 (ACE2) sequence similarities in animals, the putative receptor of this virus (Andersen et al., 2020; Li et al., 2020). Characterization of the ACE2 receptor has been focused in mammalian species that are thought to be the origin of this virus, like bats (Chiroptera), civets (Carnivora), and pangolins (Pholidota); and also focuses on some lab and domestic animals, which might work as animal models for future research (Li et al., 2020). However, other species have been let aside. So far, in experimental studies and natural cases it was shown that SARS-CoV-2 can infect some pets (dogs and cats), lab animals (primates, ferrets, Syrian hamster) and wild felids (Malayan tiger) (APHIS-USDA, 2020; Chan et al., 2020; Lu et al., 2020a; Parry, 2020; Richard et al., 2020; Shi et al., 2020; WCS, 2020). Unfortunately, the virus has settled in the human population worldwide, and while the situation is still serious, efforts are focused on protecting humankind (Ferioli et al., 2020). However, as noted, since many animals can be infected by SARS-CoV-2, its effects should no longer be studied unidirectionality in a single species, and needs to move in a more comprehensive way. To achieve this, there is a need to identify the complete range of affected hosts, not only host species of virus origin/reservoirs but also those species that can adversely be affected. There may be side effects in the ecosystem, perhaps in endangered wild animals that can be seriously impacted.
Many authors argue that humans favored this pandemic, because anthropogenic activities enhance conditions to cross the species barrier from wild animals to humans (Hassanin et al., 2020; Volpato et al., 2020). The main causes were encroaching into natural areas, the increased demand and market of bushmeat and, a synergy with globalization that promoted its rapid dissemination (Volpato et al., 2020). So, if humans triggered SARS-CoV-2 pandemic despite warnings from scientific community (Bogich et al., 2012), a new short-sightedness about the infectivity to new hosts, particularly wildlife, can boost the damage on the already dwindling ecosystems.
Given the fact that at this moment human are the main host of the SARS-CoV-2, and this virus is fully adapted to them (Cao et al., 2020), then humans may yet become viral spreaders to infect new hosts. Until 2005, adult human biomass was approximately 287 billion tons in the world (Walpole et al., 2012), that means that is the most abundant mammal species in the world. Moreover, humans populate all continents and many islands, and hence are considered ubiquitous across ecosystems; some authors even claim that there are absolutely no ecosystems free of anthropization (Sanderson et al., 2002). So, it is a fact that humans are in direct and indirect contact with wild animals, thus humans are a hazard for wildlife because of their potential for pathogen transmission (Epstein and Price, 2009). In fact, there are previous virus spillover records from human to animals (v. gr. measles, human metapneumovirus, human respiratory syncytial virus and herpes simplex virus type 1; Epstein and Price, 2009), including human coronavirus transmitted from humans to pigs in China (Chen et al., 2005) and from humans to chimpanzees (Pan troglodytes verus) in Ivory Coast (Patrono et al., 2018). Since human-influence/human-footprint occurs across all world ecosystems (Sanderson et al., 2002), the interaction with wild animals is almost impossible to avoid. Then, if SARS-CoV-2 breaks the species barrier again, from humans to wildlife, it is imperative to carry out studies on the identification of the wild species at risk, and subsequently study the impact on their populations.
For cross-species transmission in the wild, the main barriers to overcome at macro- and micro-level are: close interaction between infected and susceptible host (macro level) and virus-cell interaction by binding to surface receptors for activating cellular-signaling cascades that virus require for entry (micro level) (Marsh and Helenius, 2006). Most of our knowledge about SARS-CoV-2 comes for previous SARS-CoV research. SARS-CoV mediates the entry to the cell and fusion between the envelope and plasma membrane throughout a spike protein (S) (Decaro and Lorusso, 2020). This protein has an ectodomain divided in two subunits, S1 responsible for receptor binding and, S2 responsible for membrane fusion. The S1 subunit contains a N-terminal domain (NTD) and a C-terminal domain, for SARS-CoV the C-terminal domain contains a specific region for interaction with the host that is the receptor-binding domain (RBD). The RBD interacts specifically on a region termed receptor-binding-motif (RBM) with an ACE2, receptor for SARS-CoV. The ACE2 is formed by a domain and collectrin domain that are involved in the renin-angiotensin pathway, thereby regulating blood pressure (it is one of the most important volume regulator systems in vertebrates); in fact, ACE2 is subjected to selection pressure since deep physiological differences exist between vertebrates (Fournier et al., 2012; Li, 2013; Fam et al., 2020). The ACE2 was originally characterized as receptor of SARS-CoV, and now has also been identified as receptor for SARS-CoV-2 (Hoffmann et al., 2020; Letko et al., 2020; Ou et al., 2020). The S-protein-binding region is located in the ACE2 catalytic site (Li et al., 2006). Based on information generated for SARS-CoV, some amino acid residues at a particular position in the human ACE2 have been pointed out as an outstanding feature in the ACE2-SARS-CoV interaction (Li et al., 2020; Melin et al., 2020; Wan et al., 2020). These binding residues determine the greater (for humans) or lesser (for rodents) degree of susceptibility and are very likely to be the main drivers for cross species transmission (Wan et al., 2020). A recent study showed that ACE2 sites, located in the catalytic domain, are under positive selection, suggesting a taxonomic adaptation. Since there is a ACE2 great diversity among vertebrates, this positive selection is likely to be due to blood pressure physiology across vertebrate taxa. However, instead of this divergence among taxa, some convergence event is also noted, especially in some particular site of importance for SARS-CoV-2 recognition (Fam et al., 2020; Hajibabaei and Singer, 2020). At species level (humans), ACE2 is highly conserved, even when some amino acid sequence variations have been reported, nonetheless none of them are related with the sites of interaction between ACE2 and SARS-CoV-2 (Cao et al., 2020; Fam et al., 2020). Also, other receptors or co-receptors have been shown to enhance the SARS-CoV entry, such as L-Sing, DC-SING and L-SECtin (Jeffers et al., 2004), nevertheless, they have not been tested yet for SARS-CoV-2. Some other receptors for SARS-CoV and MERS have been tested, as Dipeptidyl Peptidase 4 (DDP4) and Aminopeptidase N (APN), but both were discarded for SARS-CoV-2 (Letko et al., 2020).
Another barrier to overcome for the virus to infect the host cell is a protease (Hoffmann et al., 2020; Letko et al., 2020). In the entry process several proteases are involved. However, one of them has been described as critical for SARS-CoV-2, the TMPRSS2, which is a protease of the Type II Transmembrane Serine Proteases (TTSP) family with a trypsin-like domain (Hoffmann et al., 2020). It is theorized that once the ACE2 receptor is binding with the S protein, the conformational change allows cleavage of TMPRSS2 with S protein, and subsequent the S protein exposes the fusion peptide that is close to the cleavage site, to finally achieve the fusion of the viral membrane with the cellular membrane (Millet and Whittaker, 2015).
The aim of this work is to identify the spillover extent of SARS-CoV-2 from humans to vertebrates by analyzing the amino acid sequences of ACE2 receptor and TMPRSS2 protease genes, and by identifying and analyzing the variant sites of the RBD and the proteolytic sites in protein S of the virus. This provides a list of vertebrate species to prioritize monitoring and protection efforts against SARS-CoV-2.
MethodsStudied speciesAll vertebrates having ACE2 (for model 1 and 2) and TMPRSS2 (for model 3) in the GenBank® were analyzed with exception of those with poor quality sequences. For models 1 and 2, 131 animal species were analyzed, 87 mammalian species (thirteen Orders), 30 birds (ten Orders), 11 reptiles (two Orders) and three amphibians (one Order). For model 3, 119 animal species were analyzed, 78 mammalian species (thirteen Orders), 29 birds (ten Orders), 10 reptiles (two Orders), and two amphibians (one Order). The best represented Orders were Primate, Carnivora and Rodentia for mammals, Passeriformes for birds, Squamata and Testudines for reptiles, and Gymnophiona for amphibians. For models 1 and 2, all sequence with good quality sequences were included. For model 3, just species having ACE2 and TMPRSS2 were included.
According with IUCN, the animals analyzed by models 1 and 2 included the following: 7 critically endangered, 13 endangered, 13 vulnerable, 4 near threatened, 75 least concern, and 2 data deficient. Additionally, 12 domestic animals were analyzed, including 11 invasive species and the remaining species are not categorized by IUCN, and are not domestic animals. The GenBank® accession numbers and the IUCN category are shown in the supplemental material (S1 Table).
Analyzed protein sequencesAngiotensin-Converting Enzyme 2 (ACE2) and Transmembrane Protease, Serine 2 (TMPRSS2).
Amino acid sequences of ACE2 and TMPRSS2 from the above describe ed animals were used with some exceptions. Sequences tagged as “Partial, Like, Predicted, Low quality,” and secondary isoforms were ruled out, except for those species with previous evidence of SARS-CoV-2 infection. Human sequence of ACE2 (ATT45083.1) and TMPRSS2 (NP001128571.1 isoform 1) were used as a pairwise comparison.
Surface glycoprotein (S) of Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).
Amino acid sequences for SARS CoV-2 protein S were obtained from GenBank® (S2 Table). The sequences were aligned and analyzed in three data matrices: (1) complete protein sequence; (2) RBD region of interaction with ACE2; and (3) Proteolytic region or recognition by the TMPRSS2 protein.
ModelsProbabilistic models were constructed based on the amino acid residue sequence for ACE2 and TMPRSS2. Two models were created for the ACE2 receptor and the third model includes the TMPRSS2. The significant residue amino acids for ACE2 that interact with the viral S protein, so far reported by Melin et al. (2020), Qiu et al. (2020), Li et al. (2020), Sun et al. (2020) and Wan et al. (2020), were used for the analysis (residues 20, 24, 30, 31, 34, 35, 37, 38, 41, 42, 53, 68, 79, 82, 83, 90, 322, 325, 329, 330, 353, 355, 357, 383, 652, 710). However, since no variation was detected, residues 355–710 were disregarded. For TMPRSS2 only the protease domain (255–492) were used for the analysis.
Model 1: similarity percentage in ACE2 Protein Binding Domain (PBD) sitesSimilarity values (proportion similar amino acid residues) were obtained from the ACE2 PBD amino acid sequences of different wild animals using human sequences as a pairwise comparison.
Model 2: physico-chemical similarity on ACE2's amino acid residuesFor each amino acid residue position a physico-chemical-similarly value (PCSV) was assigned according to physico-chemical properties of human amino acid residues and animal amino acid residues under study.
The assignation of the PCSV was as follow: four amino acid properties were used (size, hydrophobicity, charge and polarity), each one with three categories: (1) size (tiny, small and large); (2) hydrophobicity (hydrophilic, moderate, hydrophobic); (3) charge (positive, neutral, negative); and (4) polarity (polar, amphipathic, non-polar). For polarity, some amino acids are reported in two categories (i.e., amphipathic and polar or non-polar), for these cases the higher value was assigned. When human and animal showed the same amino acid residue the PCSV was assigned as 1. We assumed that an amino acid residue substitution by a different amino acid residue even sharing all the physical properties will give a 0.9 PCSV tops. Values for each physical property were assigned as follows: for each similar physical property a 0.225 was assigned; if the property was not the same but in the closest category 0.112 was assigned; and if the property was neither of those it was assigned a 0. The PCSV for each amino acid residue position was the sum of the values of the four categories. The minimum assigned value was 0.225 and the maximum was 0.9 (S3 Table). To find the total PCSV (tPCSV) for each animal species, the product of all PCSV was calculated (S4 Table).
Model 3: ACE2/TMPRSS2 co-similitudeWe construct the model by multiplying the values of model 2 by the Similarity value of the TMPRSS2. All animal species were ranked by each model.
Multiple alignments and phylogenetic analysisProtein sequences of the three genes described above were subjected to multiple alignments in 8 different data matrices: (1) ACE2 gene matrix; (2) TMPRSS2 gene matrix; (3) ACE2 PBD sites; (4) TMPRSS2 proteolytic site; (5) gene matrix ACE2 and TMPRSS2; (6) matrix genes ACE2 PBD site and TMPRSS2 proteolytic site; (7) Viral protein S PBD site; and (8) Viral protein S proteolytic site by TMPRSS2. All alignments were established using the Clustal W and Muscle algorithms included in MEGA software version 7.0.26 (Tamura et al., 2011). Phylogenetic reconstructions were performed using the eight matrices by Bayesian approximations with Mr. Bayes software version 3.2 (Huelsenbeck et al., 2001). The analysis was performed for 2 million generations with sampling trees every 100 generations. Trees with scores lower than those at the stationary phase (burn-in) were discarded, and the trees that reached the stationary phase were collected and used to build majority consensus trees.
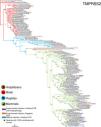
ResultsThe three models were consistent among them for most of the species, with some variation in the rank of certain particular species (Fig. 1). Model 1 gave the most different results with respect to the others. Most of the primates ranked in the first places, but interestingly, after that, carnivores, cetaceans, and wild rodents showed a relatively high potential risk. Lab rat and mouse ranked in the lowest places along with bats.
Comparative scores of the three models on different mammal species. R, Rank and M, Model. Species with infection evidence: N1 (natural, PCR negative); N3 (natural, PCR positive with no histopatological lesions); E2 (experimental; PCR positive and histopatological lesions). 1-N1 (Temmam et al., 2020); 1-E2 (Bao et al., 2020a; Lu et al., 2020a; Munster et al., 2020; Shan et al., 2020; Yu et al., 2020); 2-E2 (Lu et al., 2020a), 3-E2 (Shi et al., 2020; Halfmann et al., 2020), 4-E2 (Chan et al., 2020; Sia et al., 2020); 5-E2 (Schlottau et al., 2020; Shi et al., 2020; Kim et al., 2020), 6-E2 (Zhao et al., 2020), 7-E (Bao et al., 2020b); 1-N3 (FASFC, 2020; APHIS-USDA, 2020; Zientara, 2020); 2-N3 (WCS, 2020) and 3-N3 (Sit et al., 2020; FASFC, 2020).
Reptiles, amphibians and birds ranked below mammals, with some exception. Birds such as Empidonax traillii (Models 2 and 3), and Nothoprocta perdicaria (Model 1) classified to a level similar to that of lab rodents and bats. Reptiles with the highest rank were Chelonia mydas (Model 1), and Protobothrops mucrosquamatus (Models 2 and 3) even around some mammals such as Mus musculus and Myotis lucifugus but below Rattus norvegicus. For Models 2 and 3 most of the reptiles were at the bottom of the rank. Amphibians were at similar rank than reptiles in Model 1, but for Models 2 and 3 were around reptiles. In particular, the highest rank was Xenopus tropicalis (Model 2), and Rhinatrema bivittatum (Model 3; S5 Table).
Mammal species with natural or experimental infection evidence were usually at the top of the rank, and resilient animals were at the bottom. However, Mustela putorius did not follow this tendency.
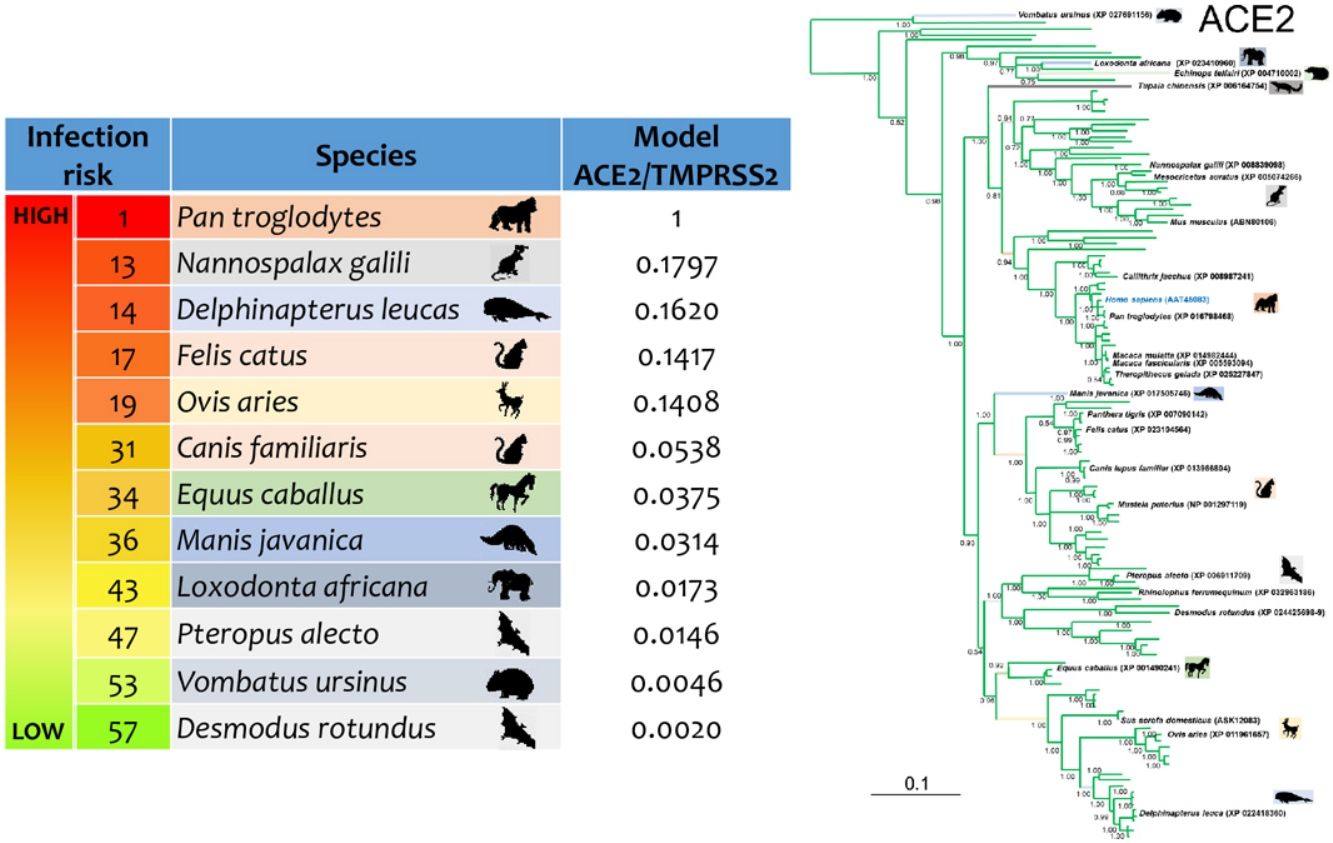
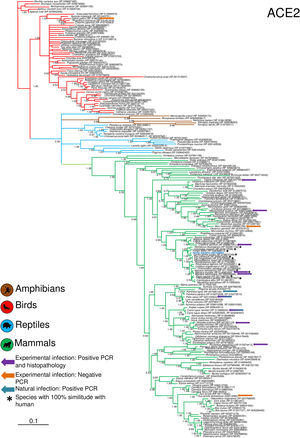
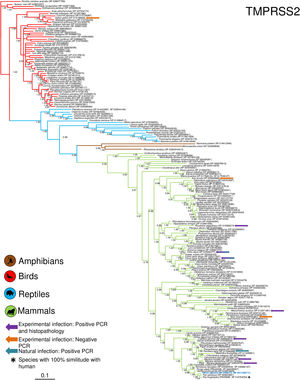
Phylogenetic analysis using the ACE2 gene showed a clear separation of the four large groups of vertebrates analyzed: amphibians, birds, reptiles and mammals (Fig. 2). Regarding the species to which the presence of SARS-CoV-2 has been identified naturally and experimentally, a high dispersion was observed throughout the tree but in clades merged with different families including: Felidae, Canidae, Mustelidae, Ursidae and Phocidae. Only on the clade for Primates fusion with other families was not observed. Similar clusters were observed when using the TMPRSS2 gene (Fig. 3) and the trees with both genes (ACE2-TMPRSS2 and ACE2 PBD motif-TMPRSS2 proteolytic site; data not shown). In the case of phylogenetic trees made with the recognition motif sequences in ACE2 and the proteolytic site in TMPRSS2, the primate clade was observed as unique in both trees, and the other families were observed with polytomies (data not shown).
Bayesian phylogenetic tree using ACE2 gene sequences for different species of domestic and wild animals. The colors on the branches indicate the vertebrate classes: amphibians (brown); birds (red); reptiles (blue) and mammals (green). The numbers in the nodes indicate the posterior probability values.
Bayesian phylogenetic tree using TMPRSS2 gene sequences for different species of domestic and wild animals. The colors on the branches indicate the different classes: amphibians (brown); birds (red); reptiles (blue) and mammals (green). The numbers in the nodes indicate the posterior probability values.
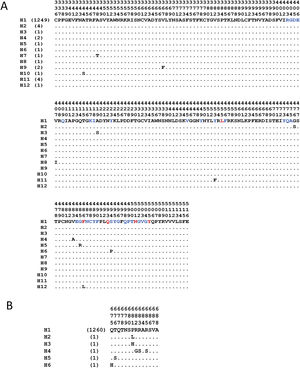
The analyses with the surface glycoprotein S showed little genetic variation, so the trees were observed with polytomies (data not show). However, both domains showed genetic variation for the RBD region, 12 haplotypes identified (Fig. 4A), with H1 being the most frequent (98.5%), and for the TMPRSS2 cut region 6 haplotypes were identified (Fig. 4B), being the most frequent H1 (99.6%). The other haplotypes presented unique frequencies.
Haplotypes of SARS-CoV-2's spike protein at the Receptor Binding Domain interaction and cleavage sites. Receptor Binding Domain for ACE2 (A) and cleavage sites by TMPRSS2 (B). The key amino acids described for the interaction with ACE2 are shown in red, and in blue other amino acid related with the interaction in SARS-CoV-2. (Lines [.]=same amino acid).
For the first time, a combination of ACE2 and TMPRSS2 was used to assess the risk of SARS-CoV-2 infection in wildlife, with similar results in previous works that only used ACE2 (Damas et al., 2020; Liu et al., 2020; Melin et al., 2020; Wu et al., 2020). All model outputs showed animal species that are, perhaps, most susceptible to SARS-CoV-2 infection, and our findings are consistent with published results of SARS-CoV-2 infection in some wildlife species (i.e. tiger, Panthera tigris), domestic animals (cats, Felis catus) and laboratory animals (ferrets, M. putorius furo, and golden Syrian hamster, Mesocricetus auratus), which were either naturally and laboratory proven (Chan et al., 2020; Shi et al., 2020; WCS, 2020), or by in silico and in vitro analysis (Damas et al., 2020; Liu et al., 2020; Melin et al., 2020; Wu et al., 2020;). The incorporation of physico-chemical properties of the amino acid residue improved the sensitivity of our models. In addition, the heterogeneous distribution of the susceptible species to natural or experimental infection in phylogenetic trees, and their association in specific clades (Felidae, Canidae, Mustelidae, Ursidae, Phocidae, and Hominidae) with other species of which the association with the virus is unknown, suggests a possible risk in nearby species.
The results indicate that Primate order is the highest risk animal group, as already pointed out in theoretical studies (Damas et al., 2020; Liu et al., 2020; Melin et al., 2020; Wu et al., 2020), and experimentally validated (Lu et al., 2020a). Then, if transmission under lab conditions is possible, some questions arise: Is transmission to primates under natural conditions feasible? Can viral infection in wild primates cause clinical disease? Some insights about what would happen came from the Human Coronavirus (HCoV)-OC43 spillover case from humans to wild chimpanzees (Patrono et al., 2018). The HCoV-OC43 belongs to the same genus as SARS-CoV-2 (Betacoronavirus), and has the potential to cause from common cold to severe respiratory illness in humans (Zhang et al., 2018). Since the end of 2016 until early 2017, an outbreak of HCoV-OC43 occurred in a chimpanzee (P. troglodytes verus) population from Taï National Park, Ivory Coast; the virus was identified in eleven out of eighteen chimpanzees, and nine of them showed signs of mild respiratory disease (Patrono et al., 2018). Though HCoV-OC43 infection in chimpanzees causes a minor illness, as in humans, its true extent in Primate order is yet unknown. However, unlike SARS-CoV, HCoV-OC43 is not considered a lethal virus (Zhang et al., 2018).
Even the present and other studies indicate that Old World primates (OWp) show greater virus-affection level than New World primates (NWp) (Melin et al., 2020), supported by the fact that NWp display poor affinity between the SARS-CoV protein S and the ACE2 (Wu et al., 2020). Actually, experimental evidence found between one species of NWp and two of OWp infected with SARS-CoV-2, showed that the former presents higher frequency of viremia than the latter, particularly 100% of six Callithrix jacchus individuals versus 44% of eighteen Macaca spp.; but they also found that viral loads and pathological lesions at microscopic level were greater in Macaca spp. than in C. jacchus (Lu et al., 2020a); also proved that NWp, at least the species C. jacchus, can get infected, excrete, and develop lesion because of SARS-CoV-2. SARS-CoV-2-infected macaques (M. fascicularis) showed virus excretion from the nose and throat in absence of clinical signs, it was detected diffuse alveolar damage foci in type I and II pneumocytes and in ciliated epithelial cells of nasal, bronchial, and bronchiolar mucosae (Rockx et al., 2020). These data show that SARS-CoV-2 causes COVID-19-like disease even in the absence of clinical signs.
Cetacean infraorder (Artiodactyla) was also identified at high risk of acquiring SARS-CoV-2. Nonetheless, until now, there is no record of infection by SARS-CoV-2 in the wild or in captivity. However, some species of this infraorder have been reported to have infections by coronaviruses of the genus Gammacoronavirus, in some cases even with clinical signs, e.g. the bottlenose dolphin coronavirus (BdCoV) and the beluga whale coronavirus SW1 (BWCoV-SW1) (Mihindukulasuriya et al., 2008; Woo et al., 2014; Schütze, 2016). The BWCoV-SW1 infection in captive-born beluga whale (Delphinapterus leucas) caused an acute liver failure, as SARS virus does, after producing a short generalized pulmonary disease (Mihindukulasuriya et al., 2008; Schütze, 2016). To date, no spillover of coronavirus has been recorded from humans to cetaceans, but the transmission of infectious agents from cetaceans to humans has been recorded (Waltzek et al., 2012). The foregoing shows that human contact with cetaceans is close enough to allow the interchange of pathogens. Actually, there are activities where zoonosis threats could happen under captivity conditions, as rescue and rehabilitation programs for marine mammals, zoos and aquarium recreational activities and animal-assisted therapies, like swim-with-dolphin programs (Hunt et al., 2008), and in wild environments like marine ecotourism, cetaceans sightings and rescue activities of stranded marine mammals (Waltzek et al., 2012). In aquatic environments, human-to-marine mammal viral transmissions has not been reported, but allegedly they have occurred from domestic animals, not in cetaceans but in seals (Carnivora: Phocidae). One of the most intriguing cases was the death (due to acute necrotizing enteritis) of three captive seals (Phoca vitulina) by an Alphacoronavirus (Bossart and Schwartz, 1990). It was hypothesized that the source of infection was feline, perhaps Feline Infectious Peritonitis virus (FIP), since feral cats were found prior to and during the outbreak and diagnostic test cross-reactivity with FIP (Bossart and Schwartz, 1990). However, the lack of molecular evidence in that case and the existence of a short coronavirus sequence from another infected harbor seal left more questions than answers, because there was no case-information but just the submitted GenBank sequence (Schütze, 2016). Interestingly, in our phylogenetic trees, these marine mammalian species share the same clade with the feline species, which also supports this hypothesis. People that inhabit or travel through the seacoast, or wastewater released by urban areas (Ahmed et al., 2020) may have been considered a risk factor for SARS CoV-2 transmission to marine mammals. Although the SARS-CoV-2 aquatic stability is unknown, other coronavirus such as SARS-CoV show good stability because they can withstand up to 14 days (Pinon and Vialette, 2018); while Murine Hepatitis Virus (MHV) up to 26 days, and Transmissible Gastroenteritis Virus (TGEV) 33 days (Casanova et al., 2009), but virions could last longer because of temperature.
The next group of animals most at risk of SARS-CoV-2 infection in our models are Carnivora. Other in silico studies using the docking approach showed that some carnivores, as Panthera pardus, P. tigris, Puma concolor, Lyxn pardinus, and Crocuta crocuta, have also less free energy, (which means more stable union, because less energy is available to make a structural change) than humans, which means more affinity for the SARS-CoV-2 S-protein (Wu et al., 2020). The carnivores susceptibility was supported on observations in cats, ferrets and dogs under experimentation (Shi et al., 2020) and under natural conditions, by the spillover from humans to animals, such as pets domestic dogs and cats (Parry, 2020), farm-raised animals American minks (Neovison vison) from Netherlands (Oreshkova et al., 2020) and Spain (MAPA, 2020), and even wild captive carnivores, such as five tigers (three Amur and two Malayan), and three African lions at the Bronx Zoo, New York, USA (WCS, 2020).
Dissent among in silico models and potential spilloverAll predictions about the infectivity of the SARS-CoV-2 came from in silico models. Based on these models and when compared with natural and experimental infections evidence, inconsistencies appear. For example, at species level, the ferret has been found to be highly susceptible under lab conditions (Shi et al., 2020), but in this and other studies, it was not shown as one of the most vulnerable species (Chan et al., 2020; Wu et al., 2020). At group level, bats and shrews (Mammalia: Soricidae) were in the same situation, because both of them rank lower in our and others models (Chan et al., 2020; Wu et al., 2020); however, under experimental conditions, bats and shrews were found receptive to infection (Schlottau et al., 2020; Zhao et al., 2020). There are explanations for this contradictory evidence. Even when ACE2 is highly divergent among vertebrate taxa, some evidence of convergence in some particular amino acid residue, important for the interaction with the SARS-CoV-2, has been documented. Under these circumstances, vertebrates ACE2 can share some particularly important amino acid residues, even in distant species from humans (Fam et al., 2020). On the other hand, because while this kind of models are useful for assessing the ability of the virus to infect the host cell through a single receptor, SARS CoV-2 might use alternative receptors to entry, like L-SING, DC-SING or L-SECtin, and different proteases (Li et al., 2006; Millet and Whittaker et al., 2015). Then, in order to improve these models, we recommend the incorporation of SARS CoV diseases markers, i.e. Interleukine 12, DC-SING, and Major Histocompatibility Complex (Chan et al., 2010; Tang et al., 2008; Keicho et al., 2009). Because the host propensity to getting affected by SARS-CoV-2 is, perhaps, not only associated to entry receptors, but to ulterior factors related to virus excretion and development of pathologies. We believe this inclusion could improve further models like the cytokines pathways (Hirano and Murakami, 2020), in addition to including such species showing susceptibility to human respiratory diseases, i.e. carnivores and primates (Enkirch and Von Messling, 2015; Patrono et al., 2018).
Another element of dissent, other than the host, is the virus per se. For example, the high variation observed in the SARS-CoV-2 RBD region (Fig. 4A), during the five months that the pandemic has lasted, made feasible the generation of new variants. These variations are non-synonymous mutations, which suggests a low pressure of natural selection that, in turn, can impact over an elevated sequence diversity in this genomic region and, consequently, may increase its hosts-species affinity. And in the case of the viral TMPRSS2 recognition region, a less genetic variation was detected, since it is represented by 6 different haplotypes that have been observed with numerous non-synonymous substitutions (Fig. 4B). However, for the viral TMPRSS2 recognition region, different repercussions are expected, such as cleavage and internalization deficiencies mediated by TMPRSS2, which might prevent the viral entry. However, since proteases are ubiquitous alternative routes could be used.
In addition to the large variation of SARS-CoV-2, some particular features make it dangerous to spillover, such as: (1) the slight difference in the ACE2 amino acid that gives recognition and forms salt bridges in SARS-CoV and SARS-CoV-2; (2) the acquisition of polybasic motif by S-SARS-CoV-2 protein and; (3) the low thermostability of S-SARS-CoV-2 protein. All these features enhance the SARS-CoV-2 adaptation to a new ACE2 and proteases system, making efficient not only the intraspecies transmission, but also the spillover to new species (Ou et al., 2020). Previous suppositions are substantiated by in vitro evidence of transfected cell with the ACE2 from different species, that have shown a broad species range (Liu et al., 2020).
All these elements highlight the possibility to spillover and, might even be drivers for a host switching and even more. Because of the high diversity of the ACE2 among mammals (Fam et al., 2020), now even it is speculated about an evolutionary race (Red Queen theory) between coronavirus (including SARS-CoV-2) and ACE2 among different hosts (Hajibabaei and Singer, 2020). However, for such scenario a tradeoff between blood pressure physiology and SARS-CoV-2 pathogenicity should be solved first.
Additional exposure and risk to diseasesIt is hypothesized that SARS-CoV-2 adapts to humans after some intraspecies transmission (Andersen et al., 2020). So, if new intraspecies transmissions occur in a different species, as has already happened in experimental infections in cats (Shi et al., 2020), the reverse spillover and virus adaptation to wildlife should happen. Moreover, recombination is an alternative mechanism that let coronavirus acquire novel biological properties in terms of virulence, host range and tissue tropism, so that coronavirus strains may increase their pathogenicity in the same species or may adapt to different species, being able to spread in the new host with exceptional swiftness (Banner and Lai, 1991).
The SARS-CoV-2 impact on new wild hosts is uncertain. To date, this virus is considered by many authors as a deadly pathogen for humans, but with varying degrees of severity (Yang et al., 2020). However, its effects on animal health are almost unknown. In humans, the severity of the disease is determined, in most cases, by patient's comorbidity status (Guan et al., 2020). However, in wild animals the disease pathophysiology and predisposing factors are unknown. As with other infectious diseases in wildlife, the pathogenic effects are driven by stressful contexts, mainly anthropogenic, as loss of habitat, climate change, loss of resources, which increase competition and pollution (Muñoz-García et al., 2018). Therefore, this stressful context in wild animals may play a similar role that comorbidity does in humans, thereby increasing the susceptibility of wild populations and, consequently, the loss of endangered species.
Once the virus enters in wild environments, social cohesion in animals could be a decisive factor for an outbreak. Then, taking individuals proximity into consideration, Primate order are at the greatest risk, not only because its receptor and protease homology, as we see in our models and phylogenetic trees, but also for its social organization and troops’ fusion-fission dynamic (Sueur et al., 2011). However, high social cohesion also exists amongst some Carnivora and many Cetacean species, even at interspecies level (Brakes, 2017; Gittleman, 1989). In this sense, we must emphasize that some scientist claim that airborne transmission is feasible through small droplets exhaled by humans, being able to move freely tens of meters from its origin (Morawska and Cao, 2020). The risk increases when social wild species are gathering in great numbers at places where people go, like national parks, and sometimes even face crowded situations (Bath and Enck, 2003).
In order to prevent person to person disease spread, during this SARS-CoV-2 global health crisis a quarantine in most of the world was imposed (Wilder-Smith and Freedman, 2020). The human containment has resulted in the appearance of wild animals in some cities around world, and their presence indicates that humans and wildlife are closer than ever (Lewis, 2020). This proximity exists even in natural areas, where ecotourism has become one of the major economic activities, owing to wildlife observations (Zhang and Zhang, 2020). Although at the moment ecotourism has collapsed by SARS-CoV-2 quarantine, it will be reactivated in the near future (Karabulut et al., 2020), and go back to be an important activity for interaction with wildlife that looming new spillovers and virus dissemination. On 2018, the world tourism organization estimates around 1407 million of international tourism arrivals (UNWTO, 2019). Ecotourism hotspots are mainly located at Central America, Caribbean, southeast Africa and south Asia (Smith, 2019). Unfortunately, ecotourist hotspots frequently match with endangered ecological regions, as those located in Brazil, Mexico and Costa Rica, countries with high endemism and diversity of animal species (Elbers, 2011).
But not just natural areas are at risk. Wildlife may become infected through drinking water from natural bodies contaminated with sewage, mainly from hospitals and medical centers, or by ingestion of mollusks or other aquatic organisms that bioaccumulate viruses from the aquatic environment (Franklin and Bevins, 2020). Since SARS-CoV-2 was identified in human stool samples (Xu et al., 2020), then the waterborne route cannot be ruled out.
ConclusionsBecause viral dissemination is imminent, we encourage everyone involved, such as wildlife workers, veterinarians, and biologists, to develop surveillance and impact monitoring for preventing possible devastating impacts. Since this is the first time we try to evaluate a global disease from an ecological perspective, anticipating SARS-CoV-2 will affect an extensive mammal taxon, more detailed models with new data should be generated as soon as possible towards avoiding wildlife species extinctions.
Some pieces of the puzzle are still missing, but policies and decisions should be taken. We strongly recommend following four main guidelines:
- 1.
To urgently carry out wildlife monitoring for SARS-CoV-2 detection, by active (trapping and sampling) and passive surveillance (sampling animals opportunistically, like those who have been roadkilled, hunted or arrived to rescue centers), and even include citizen reports.
- 2.
To avoid or limit human direct contact with wildlife, particular emphasis should be placed on nature reserves. Cautions should be exercised by wildlife workers, researchers, veterinarians, and biologist before coming into contact with animals, as in performing diagnostic tests, and to use personal protective equipment.
- 3.
Appropriate waste disposal must be carried out, especially with sewage from hospitals and medical centers, to avoid environmental pollution.
- 4.
Emergency protocols for addressing a wildlife outbreak should be immediately developed.
During the review of this article one particularly important document was developed by collaboration of many organization OIE, IUCN, SSC, and WHSG, “Guidelines for Working with Free-Ranging Wild Mammals in the Era of the COVID-19 Pandemic.” This guide is aiming to prevent the transmission to wild mammals, highlighting the exposure risk (contact exposure, aerosol exposure, and environmental exposure), and some mitigation strategies (minimize, assess, and protect). At the same time this document provides some additional resource for specific groups such as great apes, bats, felid, and small carnivores (OIE, 2020). We encourage to review this document as a first step in the strategy to limit SARS-CoV-2 spread among wildlife. However, we should keep in mind that transmission could occur, and we still need protocols for monitoring and mitigation strategies once SARS-CoV-2 is detected.
FundingThis research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
ERF wants to thank Prof. Emmanuel Do Linh San from University of Fort Hare and the African Small Carnivore Research Initiatives (ASCaRIs https://ascaris.org) for hosting him during the sabbatical stay.